Featured Publications
Get to know some of the latest publications by our faculties.
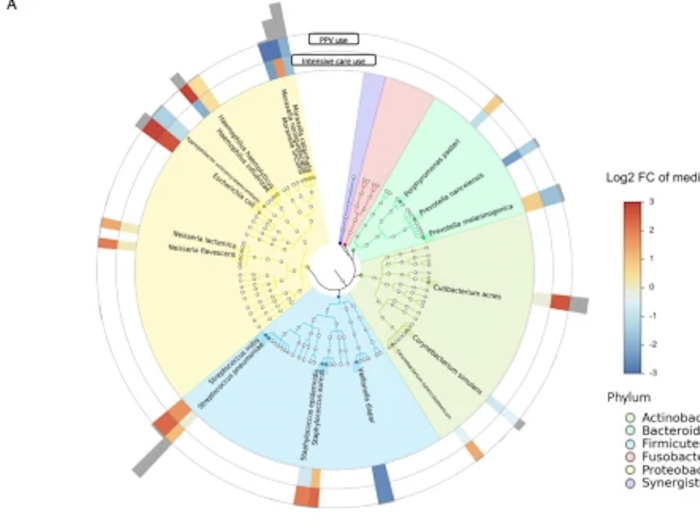
Integrated relationship of nasopharyngeal airway host response and microbiome associates with bronchiolitis severity
January 7, 2026
Bronchiolitis is a leading cause of infant hospitalizations but its immunopathology remains poorly understood.
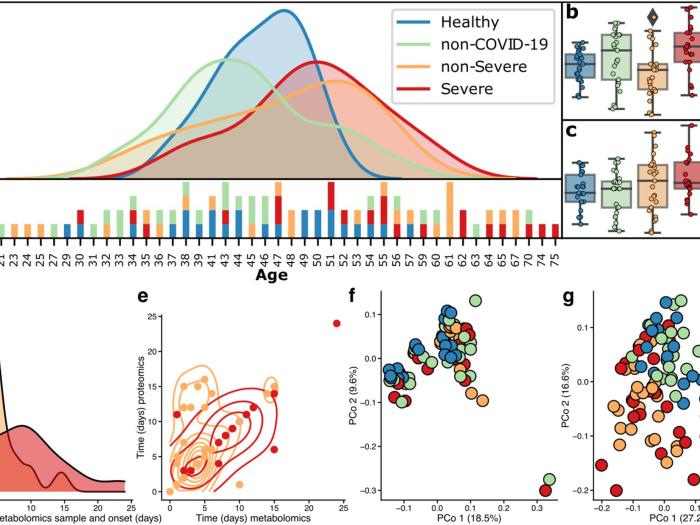
Metabolite, protein, and tissue dysfunction associated with COVID‑19 disease severity
January 7, 2026
Proteins are direct products of the genome and metabolites are functional products of interactions between the host and other factors.
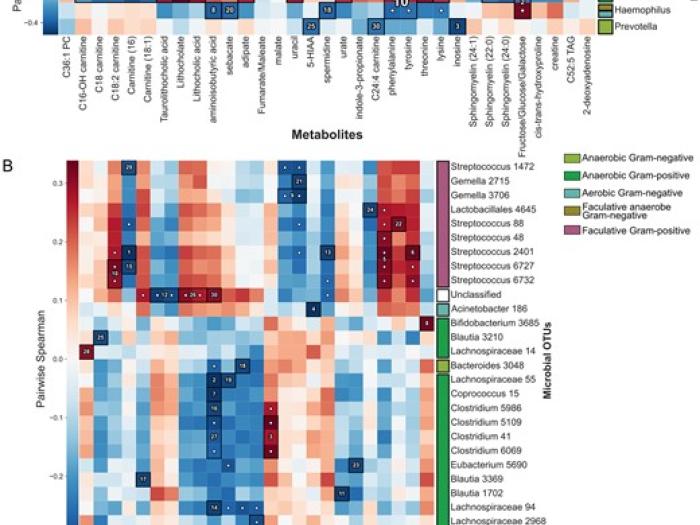
High-sensitivity pattern discovery in large, paired multiomic datasets
January 7, 2026
We present a novel framework which integrates hierarchical hypothesis testing with FDR correction to reveal linear and non-linear relationships among data.
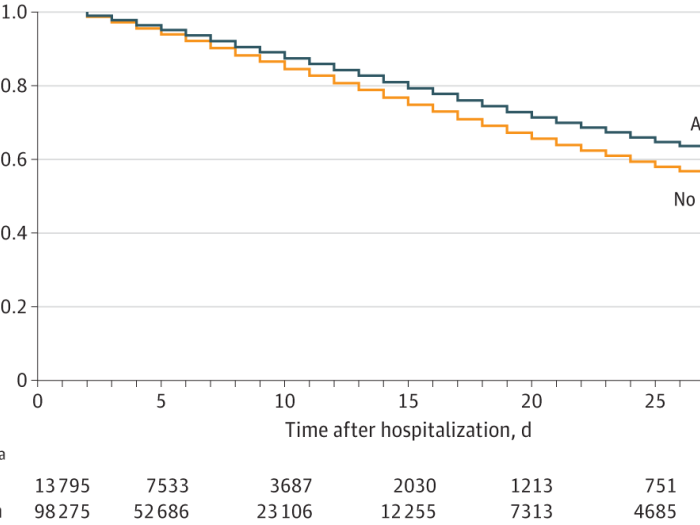
Association of Early Aspirin Use With In-Hospital Mortality in Patients With Moderate COVID-19
January 7, 2026
Prior studies suggest aspirin may be associated with reduced mortality in high-risk patients with COVID-19, but patients with moderate COVID-19 are not studied.

